променени са 61 файла, в които са добавени 266 реда и са изтрити 460 реда
BIN
MRI_volumes_customtemplate_float32/Inf_NaN_stableAD__I11767_masked_brain.nii.nii
BIN
MRI_volumes_customtemplate_float32/Inf_NaN_stableAD__I11879_masked_brain.nii.nii
BIN
MRI_volumes_customtemplate_float32/Inf_NaN_stableAD__I12061_masked_brain.nii.nii
BIN
MRI_volumes_customtemplate_float32/Inf_NaN_stableAD__I13509_masked_brain.nii.nii
BIN
MRI_volumes_customtemplate_float32/Inf_NaN_stableAD__I13807_masked_brain.nii.nii
BIN
MRI_volumes_customtemplate_float32/Inf_NaN_stableAD__I14166_masked_brain.nii.nii
BIN
MRI_volumes_customtemplate_float32/Inf_NaN_stableAD__I14808_masked_brain.nii.nii
BIN
MRI_volumes_customtemplate_float32/Inf_NaN_stableAD__I16169_masked_brain.nii.nii
BIN
MRI_volumes_customtemplate_float32/Inf_NaN_stableAD__I16238_masked_brain.nii.nii
BIN
MRI_volumes_customtemplate_float32/Inf_NaN_stableAD__I16740_masked_brain.nii.nii
BIN
MRI_volumes_customtemplate_float32/Inf_NaN_stableAD__I16828_masked_brain.nii.nii
BIN
MRI_volumes_customtemplate_float32/Inf_NaN_stableAD__I17363_masked_brain.nii.nii
BIN
MRI_volumes_customtemplate_float32/Inf_NaN_stableAD__I17415_masked_brain.nii.nii
BIN
MRI_volumes_customtemplate_float32/Inf_NaN_stableAD__I17585_masked_brain.nii.nii
BIN
MRI_volumes_customtemplate_float32/Inf_NaN_stableAD__I20080_masked_brain.nii.nii
BIN
MRI_volumes_customtemplate_float32/Inf_NaN_stableAD__I20332_masked_brain.nii.nii
BIN
MRI_volumes_customtemplate_float32/Inf_NaN_stableAD__I20506_masked_brain.nii.nii
BIN
MRI_volumes_customtemplate_float32/Inf_NaN_stableAD__I20771_masked_brain.nii.nii
BIN
MRI_volumes_customtemplate_float32/Inf_NaN_stableAD__I23153_masked_brain.nii.nii
BIN
MRI_volumes_customtemplate_float32/Inf_NaN_stableAD__I23431_masked_brain.nii.nii
BIN
MRI_volumes_customtemplate_float32/Inf_NaN_stableNL__I11771_masked_brain.nii.nii
BIN
MRI_volumes_customtemplate_float32/Inf_NaN_stableNL__I12151_masked_brain.nii.nii
BIN
MRI_volumes_customtemplate_float32/Inf_NaN_stableNL__I13447_masked_brain.nii.nii
BIN
MRI_volumes_customtemplate_float32/Inf_NaN_stableNL__I14114_masked_brain.nii.nii
BIN
MRI_volumes_customtemplate_float32/Inf_NaN_stableNL__I14783_masked_brain.nii.nii
BIN
MRI_volumes_customtemplate_float32/Inf_NaN_stableNL__I16867_masked_brain.nii.nii
BIN
MRI_volumes_customtemplate_float32/Inf_NaN_stableNL__I17508_masked_brain.nii.nii
BIN
MRI_volumes_customtemplate_float32/Inf_NaN_stableNL__I18931_masked_brain.nii.nii
BIN
MRI_volumes_customtemplate_float32/Inf_NaN_stableNL__I20550_masked_brain.nii.nii
BIN
MRI_volumes_customtemplate_float32/Inf_NaN_stableNL__I20726_masked_brain.nii.nii
BIN
MRI_volumes_customtemplate_float32/Inf_NaN_stableNL__I23089_masked_brain.nii.nii
BIN
MRI_volumes_customtemplate_float32/Inf_NaN_stableNL__I23667_masked_brain.nii.nii
BIN
MRI_volumes_customtemplate_float32/Inf_NaN_stableNL__I23798_masked_brain.nii.nii
BIN
MRI_volumes_customtemplate_float32/Inf_NaN_stableNL__I23901_masked_brain.nii.nii
BIN
MRI_volumes_customtemplate_float32/Inf_NaN_stableNL__I24641_masked_brain.nii.nii
BIN
MRI_volumes_customtemplate_float32/Inf_NaN_stableNL__I25715_masked_brain.nii.nii
BIN
MRI_volumes_customtemplate_float32/Inf_NaN_stableNL__I26030_masked_brain.nii.nii
BIN
MRI_volumes_customtemplate_float32/Inf_NaN_stableNL__I26940_masked_brain.nii.nii
BIN
MRI_volumes_customtemplate_float32/Inf_NaN_stableNL__I26947_masked_brain.nii.nii
BIN
MRI_volumes_customtemplate_float32/Inf_NaN_stableNL__I28549_masked_brain.nii.nii
BIN
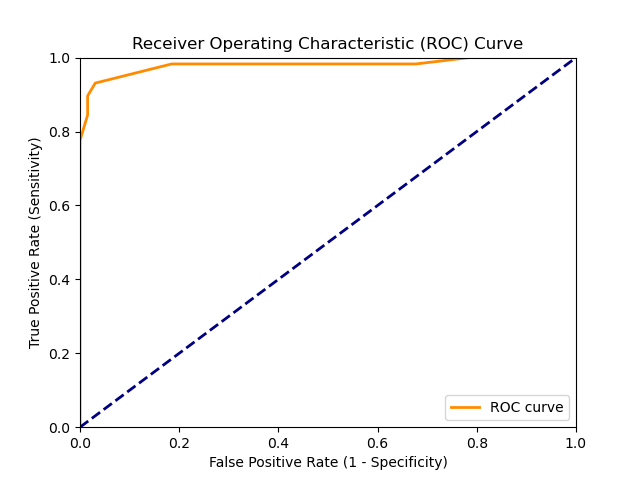
ROC.png
BIN
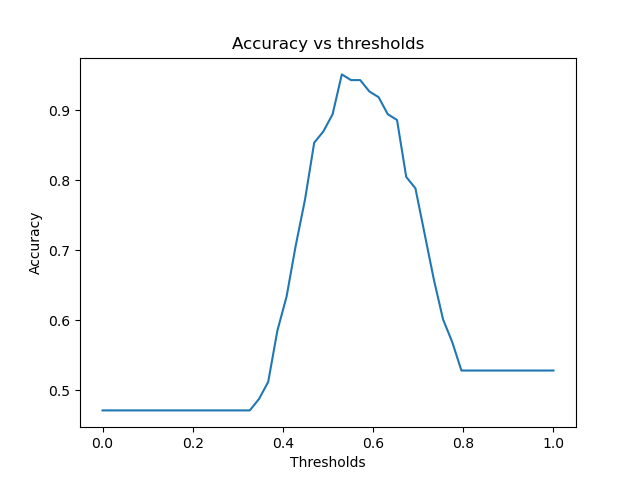
acc.png
BIN
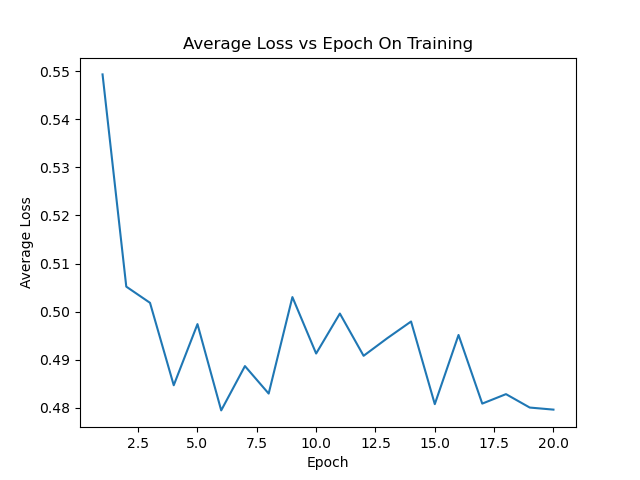
avgloss_epoch_curve.png
BIN
cnn_net.pth
+ 21
- 0
cnn_net_data.csv
|
|||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
BIN
first_train_cnn.pth
+ 10
- 20
main.py
|
|||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
+ 3
- 3
original_model/mci_train.py
|
|||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
+ 0
- 0
original_model/utils/__init__.py → original_model/utils_old/__init__.py
+ 0
- 0
original_model/utils/augmentation.py → original_model/utils_old/augmentation.py
+ 0
- 0
original_model/utils/heatmapPlotting.py → original_model/utils_old/heatmapPlotting.py
+ 0
- 0
original_model/utils/models.py → original_model/utils_old/models.py
+ 0
- 0
original_model/utils/patientsort.py → original_model/utils_old/patientsort.py
+ 0
- 0
original_model/utils/preprocess.py → original_model/utils_old/preprocess.py
+ 0
- 0
original_model/utils/sepconv3D.py → original_model/utils_old/sepconv3D.py
+ 228
- 0
utils/CNN.py
|
|||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
+ 4
- 1
utils/newCNN_Layers.py → utils/CNN_Layers.py
|
|||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
+ 0
- 78
utils/CNN_methods.py
|
|||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
+ 0
- 222
utils/models.py
|
|||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
+ 0
- 136
utils/newCNN.py
|
|||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
BIN
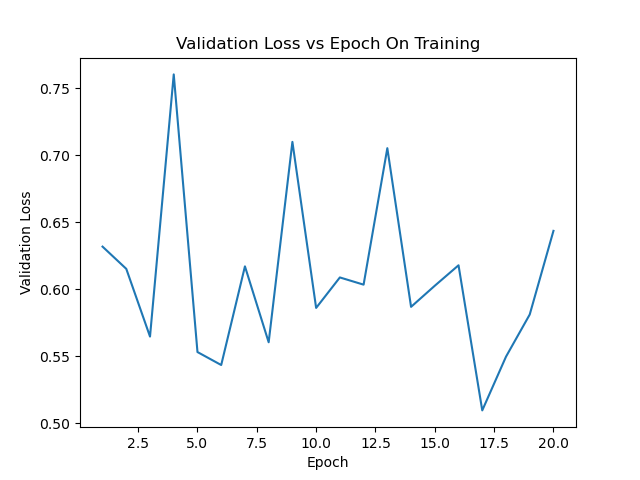
valloss_epoch_curve.png