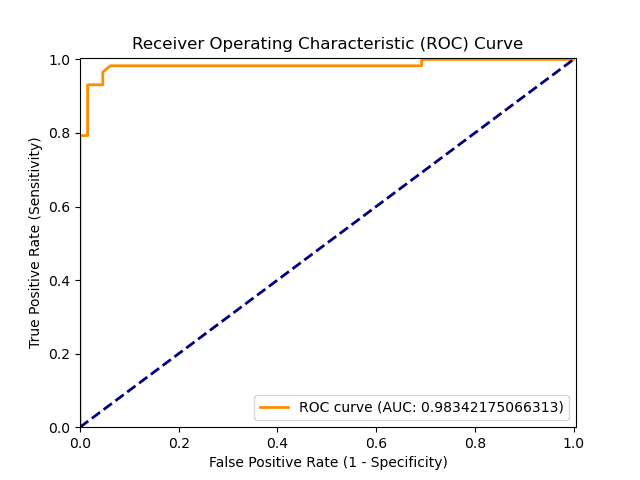
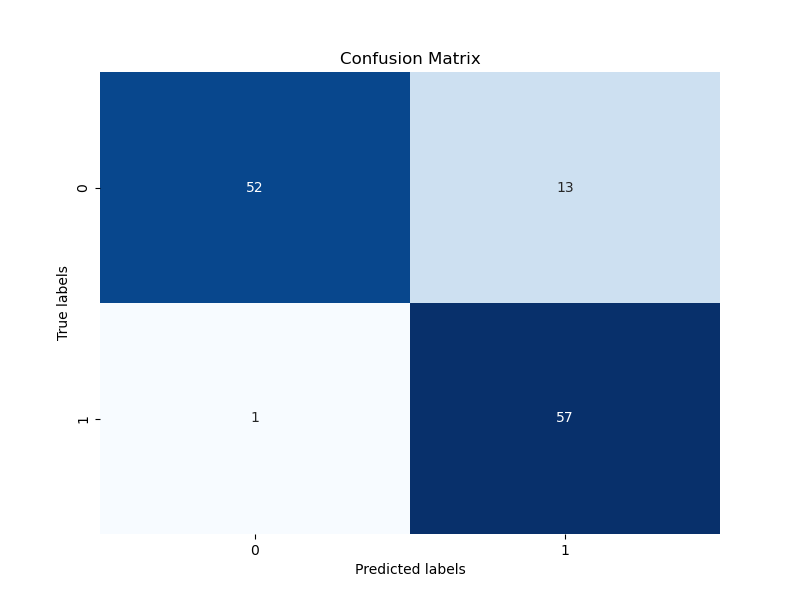
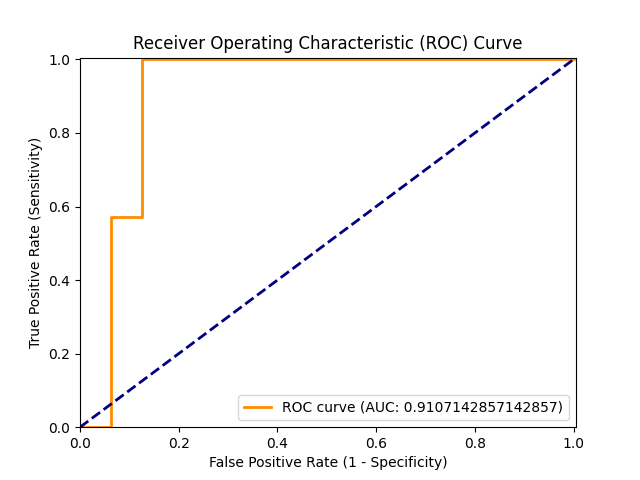
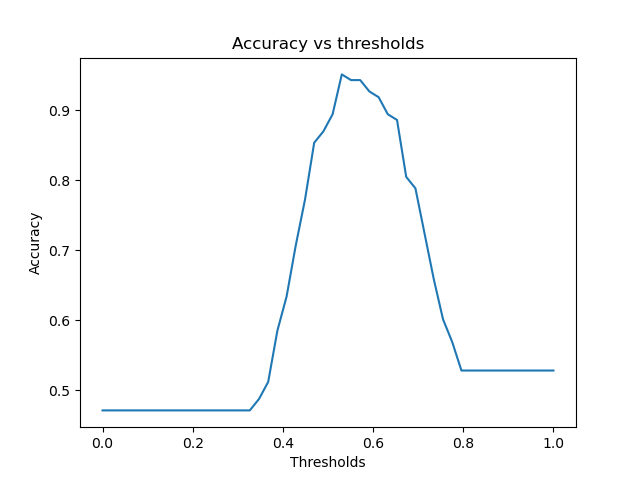
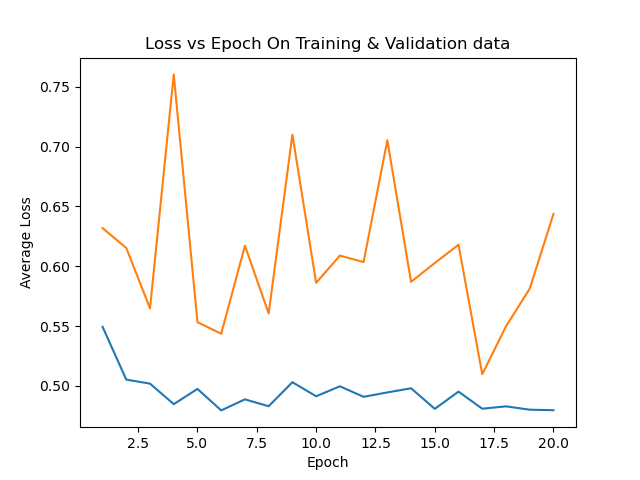
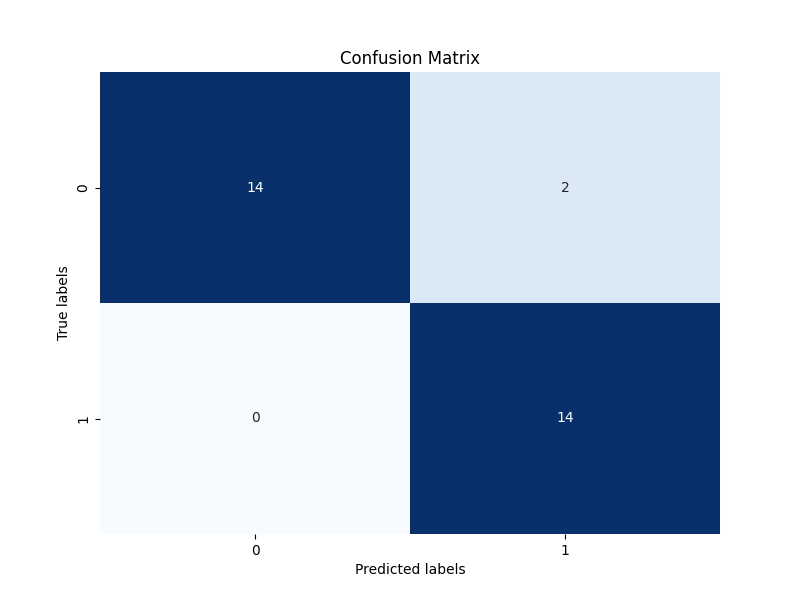
|
|
@@ -1,13 +1,16 @@
|
|
|
import os
|
|
|
import torch
|
|
|
+from utils.train_methods import train, evaluate
|
|
|
from utils.CNN import CNN_Net
|
|
|
from torch import nn
|
|
|
from torch.utils.data import DataLoader, ConcatDataset
|
|
|
from torchvision import transforms
|
|
|
from sklearn.model_selection import KFold, StratifiedKFold
|
|
|
+from sklearn.metrics import roc_curve, auc, confusion_matrix, classification_report
|
|
|
from utils.preprocess import prepare_datasets, prepare_predict
|
|
|
import numpy as np
|
|
|
import matplotlib.pyplot as plt
|
|
|
+import time
|
|
|
|
|
|
|
|
|
def reset_weights(m):
|
|
|
@@ -20,12 +23,20 @@ def reset_weights(m):
|
|
|
print(f'Reset trainable parameters of layer = {layer}')
|
|
|
layer.reset_parameters()
|
|
|
|
|
|
+
|
|
|
if __name__ == '__main__':
|
|
|
|
|
|
- # Might have to replace datapaths or separate between training and testing
|
|
|
+ print("--- RUNNING K-FOLD ---")
|
|
|
+ print("Pytorch Version: " + torch.__version__)
|
|
|
+ current_time = time.localtime()
|
|
|
+ print(time.strftime("%Y-%m-%d_%H:%M", current_time))
|
|
|
+
|
|
|
+ # might have to replace datapaths or separate between training and testing
|
|
|
model_filepath = '/data/data_wnx1/rschuurs/Pytorch_CNN-RNN'
|
|
|
CNN_filepath = '/data/data_wnx1/rschuurs/Pytorch_CNN-RNN/cnn_net.pth' # cnn_net.pth
|
|
|
+ # small dataset
|
|
|
# mri_datapath = '/data/data_wnx1/rschuurs/Pytorch_CNN-RNN/PET_volumes_customtemplate_float32/' # Small Test
|
|
|
+ # big dataset
|
|
|
mri_datapath = '/data/data_wnx1/_Data/AlzheimersDL/CNN+RNN-2class-1cnn+data/PET_volumes_customtemplate_float32/' # Real data
|
|
|
annotations_datapath = './data/data_wnx1/rschuurs/Pytorch_CNN-RNN/LP_ADNIMERGE.csv'
|
|
|
|
|
|
@@ -40,15 +51,15 @@ if __name__ == '__main__':
|
|
|
}
|
|
|
|
|
|
# Configuration options
|
|
|
- k_folds = 5 # TODO
|
|
|
- num_epochs = 1
|
|
|
+ k_folds = 5
|
|
|
+ num_epochs = 10
|
|
|
loss_function = nn.CrossEntropyLoss()
|
|
|
|
|
|
# For fold results
|
|
|
results = {}
|
|
|
|
|
|
# Set fixed random number seed
|
|
|
- torch.manual_seed(42)
|
|
|
+ torch.manual_seed(42) # todo
|
|
|
|
|
|
training_data, val_data, test_data = prepare_datasets(mri_datapath, val_split=0.2, seed=12)
|
|
|
|
|
|
@@ -79,49 +90,13 @@ if __name__ == '__main__':
|
|
|
dataset,
|
|
|
batch_size=10, sampler=test_subsampler)
|
|
|
|
|
|
- # Init the neural network
|
|
|
- network = CNN_Net(prps=properties, final_layer_size=2)
|
|
|
- network.apply(reset_weights)
|
|
|
-
|
|
|
- # Initialize optimizer
|
|
|
- optimizer = torch.optim.Adam(network.parameters(), lr=1e-5)
|
|
|
+ # Init the neural model
|
|
|
+ model = CNN_Net(prps=properties, final_layer_size=2)
|
|
|
+ model.apply(reset_weights)
|
|
|
+ model.cuda()
|
|
|
|
|
|
# Run the training loop for defined number of epochs
|
|
|
- for epoch in range(0, num_epochs):
|
|
|
-
|
|
|
- # Print epoch
|
|
|
- print(f'Starting epoch {epoch + 1}')
|
|
|
-
|
|
|
- # Set current loss value
|
|
|
- current_loss = 0.0
|
|
|
-
|
|
|
- # Iterate over the DataLoader for training data
|
|
|
- for i, data in enumerate(trainloader, 0):
|
|
|
-
|
|
|
- # Get inputs
|
|
|
- inputs, targets = data
|
|
|
-
|
|
|
- # Zero the gradients
|
|
|
- optimizer.zero_grad()
|
|
|
-
|
|
|
- # Perform forward pass
|
|
|
- outputs = network(inputs)
|
|
|
-
|
|
|
- # Compute loss
|
|
|
- loss = loss_function(outputs, targets)
|
|
|
-
|
|
|
- # Perform backward pass
|
|
|
- loss.backward()
|
|
|
-
|
|
|
- # Perform optimization
|
|
|
- optimizer.step()
|
|
|
-
|
|
|
- # Print statistics
|
|
|
- current_loss += loss.item()
|
|
|
- if i % 500 == 499:
|
|
|
- print('Loss after mini-batch %5d: %.3f' %
|
|
|
- (i + 1, current_loss / 500))
|
|
|
- current_loss = 0.0
|
|
|
+ train(model, trainloader, testloader, CNN_filepath, epochs=num_epochs, graphs=True)
|
|
|
|
|
|
# Process is complete.
|
|
|
print('Training process has finished. Saving trained model.')
|
|
|
@@ -131,81 +106,31 @@ if __name__ == '__main__':
|
|
|
|
|
|
# Saving the model
|
|
|
save_path = f'./model-fold-{fold}.pth'
|
|
|
- torch.save(network.state_dict(), save_path)
|
|
|
+ torch.save(model.state_dict(), save_path)
|
|
|
|
|
|
# Evaluation for this fold
|
|
|
- correct, total = 0, 0
|
|
|
- with torch.no_grad():
|
|
|
-
|
|
|
- predictions = []
|
|
|
- true_labels = []
|
|
|
-
|
|
|
- # Iterate over the test data and generate predictions
|
|
|
- for i, data in enumerate(testloader, 0):
|
|
|
- # Get inputs
|
|
|
- inputs, targets = data
|
|
|
+ results = evaluate(model, testloader, graphs=True, k_folds=k_folds, fold=fold, results=results)
|
|
|
|
|
|
- # Generate outputs
|
|
|
- outputs = network(inputs)
|
|
|
|
|
|
- # Set total and correct
|
|
|
- _, predicted = torch.max(outputs.data, 1)
|
|
|
- total += targets.size(0)
|
|
|
- correct += (predicted == targets).sum().item()
|
|
|
-
|
|
|
- predictions.extend(outputs.data[:, 1].cpu().numpy()) # Grabs probability of positive
|
|
|
- true_labels.extend(targets.cpu().numpy())
|
|
|
-
|
|
|
- # Print accuracy
|
|
|
- print('Accuracy for fold %d: %d %%' % (fold, 100.0 * correct / total))
|
|
|
- print('--------------------------------')
|
|
|
- results[fold] = 100.0 * (correct / total)
|
|
|
-
|
|
|
-
|
|
|
- # MAKES ROC CURVE
|
|
|
- thresholds = np.linspace(0, 1, num=50)
|
|
|
- tpr = []
|
|
|
- fpr = []
|
|
|
- acc = []
|
|
|
-
|
|
|
- true_labels = np.array(true_labels)
|
|
|
-
|
|
|
- for threshold in thresholds:
|
|
|
- # Thresholding the predictions (meaning all predictions above threshold are considered positive)
|
|
|
- thresholded_predictions = (predictions >= threshold).astype(int)
|
|
|
-
|
|
|
- # Calculating true positives, false positives, true negatives, false negatives
|
|
|
- true_positives = np.sum((thresholded_predictions == 1) & (true_labels == 1))
|
|
|
- false_positives = np.sum((thresholded_predictions == 1) & (true_labels == 0))
|
|
|
- true_negatives = np.sum((thresholded_predictions == 0) & (true_labels == 0))
|
|
|
- false_negatives = np.sum((thresholded_predictions == 0) & (true_labels == 1))
|
|
|
-
|
|
|
- accuracy = (true_positives + true_negatives) / (
|
|
|
- true_positives + false_positives + true_negatives + false_negatives)
|
|
|
-
|
|
|
- # Calculate TPR and FPR
|
|
|
- tpr.append(true_positives / (true_positives + false_negatives))
|
|
|
- fpr.append(false_positives / (false_positives + true_negatives))
|
|
|
- acc.append(accuracy)
|
|
|
-
|
|
|
- plt.plot(fpr, tpr, lw=2, label=f'ROC Fold {fold}')
|
|
|
- plt.plot([0, 1], [0, 1], color='navy', lw=2, linestyle='--')
|
|
|
- plt.xlim([0.0, 1.0])
|
|
|
- plt.ylim([0.0, 1.0])
|
|
|
-
|
|
|
- plt.xlabel('False Positive Rate (1 - Specificity)')
|
|
|
- plt.ylabel('True Positive Rate (Sensitivity)')
|
|
|
- plt.title('Receiver Operating Characteristic (ROC) Curve')
|
|
|
- plt.legend(loc="lower right")
|
|
|
-
|
|
|
- plt.savefig(f'./ROC_{k_folds}_Folds.png')
|
|
|
- plt.show()
|
|
|
-
|
|
|
- # Print fold results
|
|
|
- print(f'K-FOLD CROSS VALIDATION RESULTS FOR {k_folds} FOLDS')
|
|
|
- print('--------------------------------')
|
|
|
- sum = 0.0
|
|
|
- for key, value in results.items():
|
|
|
- print(f'Fold {key}: {value} %')
|
|
|
- sum += value
|
|
|
- print(f'Average: {sum / len(results.items())} %')
|
|
|
+ # Print fold results
|
|
|
+ print(f'K-FOLD CROSS VALIDATION RESULTS FOR {k_folds} FOLDS')
|
|
|
+ print('--------------------------------')
|
|
|
+ sum = 0.0
|
|
|
+ for key, value in results.items():
|
|
|
+ print(f'Fold {key}: {value} %')
|
|
|
+ sum += value
|
|
|
+ print(f'Average: {sum / len(results.items())} %')
|
|
|
+
|
|
|
+ # Saves to .txt if last one
|
|
|
+ if(fold==k_folds-1):
|
|
|
+ time_string = time.strftime("%Y-%m-%d_%H:%M", current_time)
|
|
|
+ txt = open(f"{k_folds}_folds_{time_string}.txt", "w")
|
|
|
+ txt.write('--------------------------------')
|
|
|
+ txt.write(f'K-FOLD CROSS VALIDATION RESULTS FOR {k_folds} FOLDS')
|
|
|
+ txt.write('--------------------------------')
|
|
|
+ sum = 0.0
|
|
|
+ for key, value in results.items():
|
|
|
+ txt.write(f'Fold {key}: {value} %')
|
|
|
+ sum += value
|
|
|
+ txt.write(f'Average: {sum / len(results.items())} %')
|
|
|
+ txt.close()
|