31 changed files with 92 additions and 29 deletions
BIN
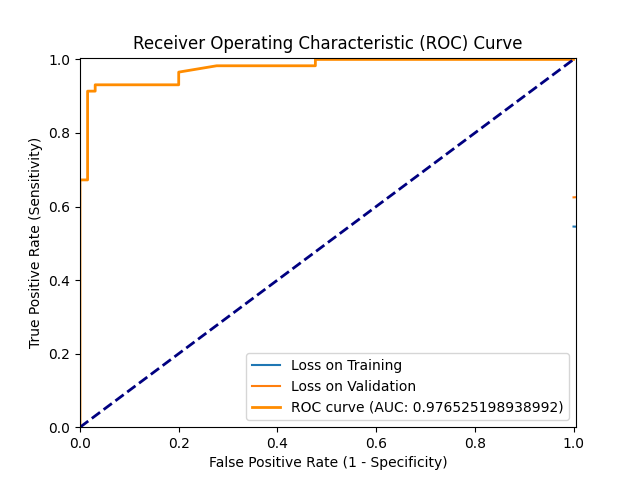
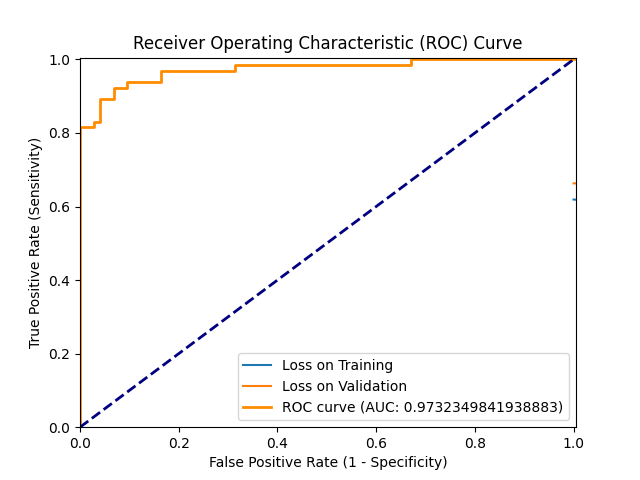
ROC_2024-08-13_14.36.png
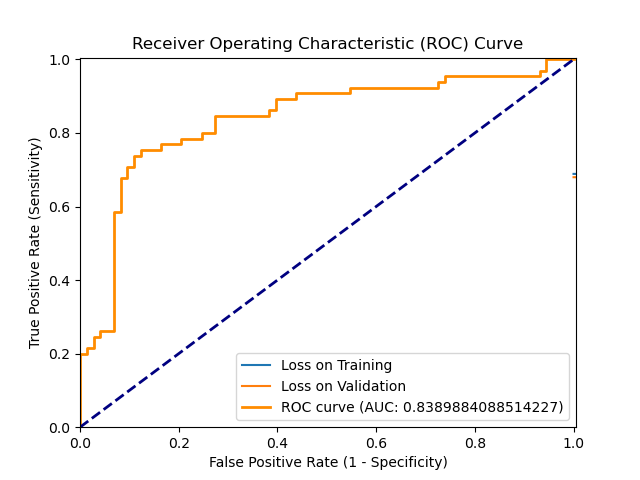
BIN
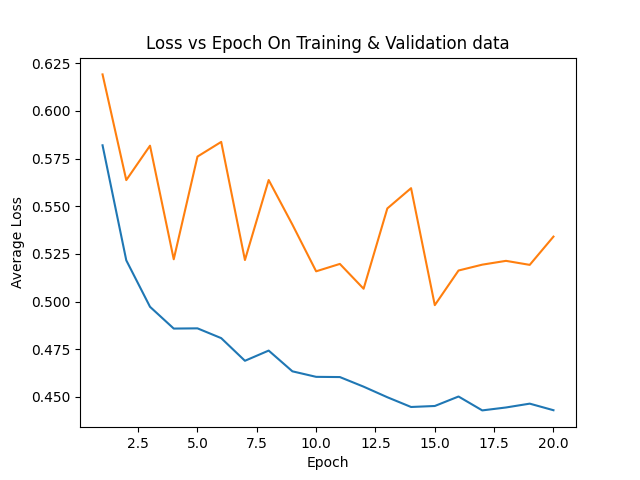
avgloss_epoch_curve_2024-08-13_14.36.png
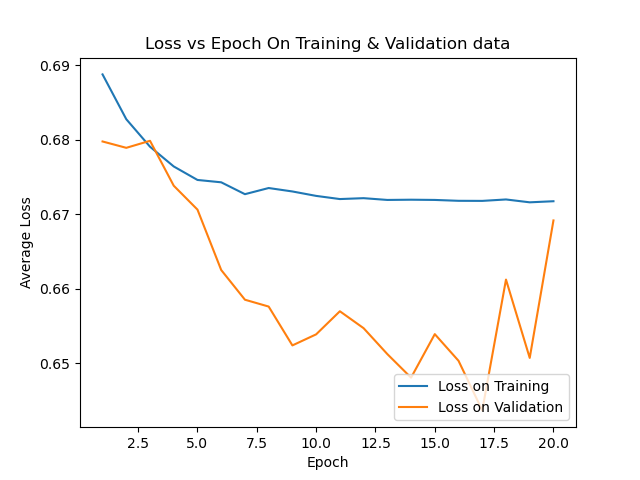
BIN
cnn_net.pth
+ 21
- 0
cnn_net_data_2024-08-13_14.36.csv
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
BIN
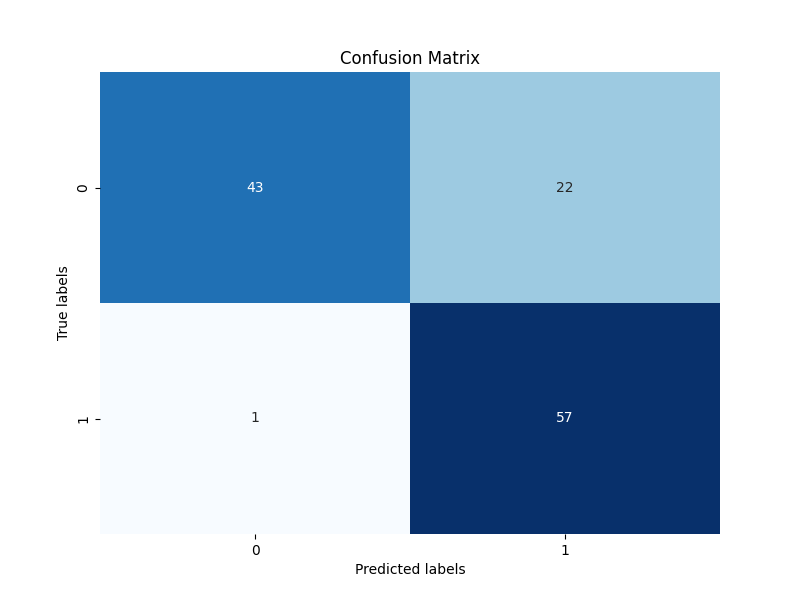
confusion_matrix_2024-08-13_14.36.png
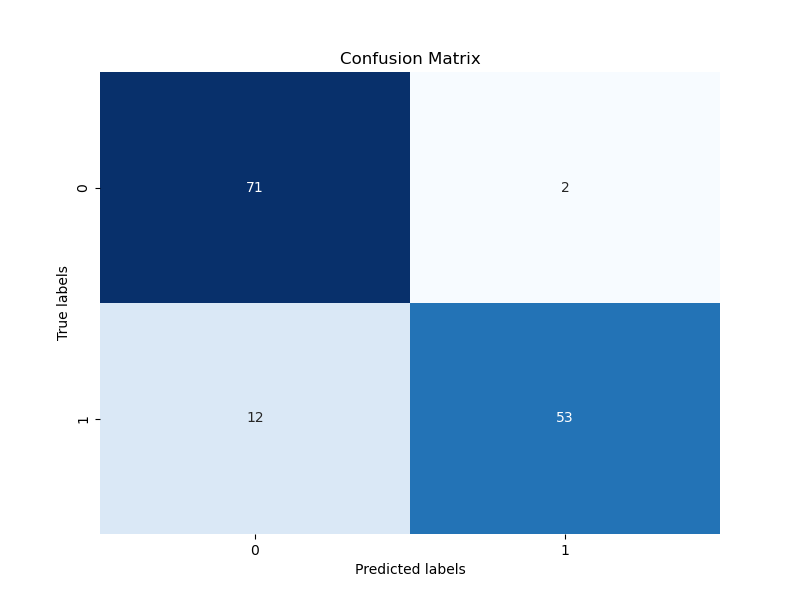
+ 0
- 0
figures/5-fold_7_26/5_folds_2024-07-26_14:26.txt → figures/5-fold_7_26/5_folds_2024-07-26_14.26.txt
+ 0
- 0
figures/5-fold_7_26/ROC_5_Folds_2024-07-26_15:22.png → figures/5-fold_7_26/ROC_5_Folds_2024-07-26_15.22.png

+ 0
- 0
figures/5-fold_7_26/cnn_net_data_2024-07-26_14:37.csv → figures/5-fold_7_26/cnn_net_data_2024-07-26_14.37.csv
+ 0
- 0
figures/5-fold_7_26/cnn_net_data_2024-07-26_14:48.csv → figures/5-fold_7_26/cnn_net_data_2024-07-26_14.48.csv
+ 0
- 0
figures/5-fold_7_26/cnn_net_data_2024-07-26_15:00.csv → figures/5-fold_7_26/cnn_net_data_2024-07-26_15.00.csv
+ 0
- 0
figures/5-fold_7_26/cnn_net_data_2024-07-26_15:11.csv → figures/5-fold_7_26/cnn_net_data_2024-07-26_15.11.csv
+ 0
- 0
figures/5-fold_7_26/cnn_net_data_2024-07-26_15:22.csv → figures/5-fold_7_26/cnn_net_data_2024-07-26_15.22.csv
+ 0
- 0
figures/Age_and_Sex_Data_Run/ROC_2024-05-30_15:48.png → figures/Age_and_Sex_Data_Run/ROC_2024-05-30_15.48.png
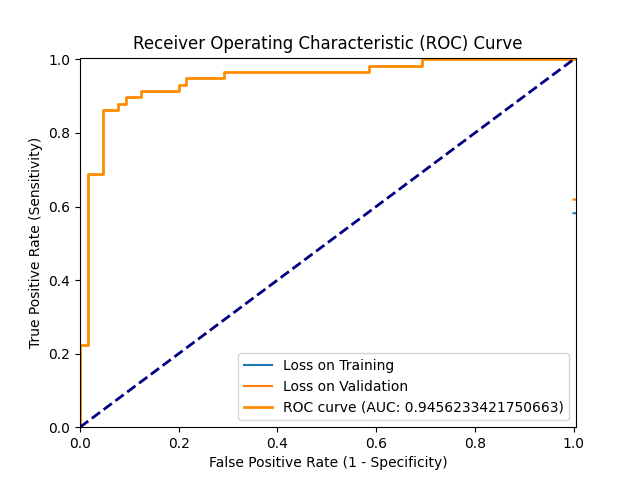
+ 0
- 0
figures/Age_and_Sex_Data_Run/avgloss_epoch_curve_2024-05-30_15:48.png → figures/Age_and_Sex_Data_Run/avgloss_epoch_curve_2024-05-30_15.48.png
+ 0
- 0
figures/Age_and_Sex_Data_Run/cnn_net_data_2024-05-30_15:48.csv → figures/Age_and_Sex_Data_Run/cnn_net_data_2024-05-30_15.48.csv
+ 0
- 0
figures/Age_and_Sex_Data_Run/confusion_matrix_2024-05-30_15:48.png → figures/Age_and_Sex_Data_Run/confusion_matrix_2024-05-30_15.48.png

+ 0
- 0
figures/Pre-Clinical_Data_Run/ROC_2024-05-30_11:39.png → figures/Pre-Clinical_Data_Run/ROC_2024-05-30_11.39.png
+ 0
- 0
figures/Pre-Clinical_Data_Run/avgloss_epoch_curve_2024-05-30_11:39.png → figures/Pre-Clinical_Data_Run/avgloss_epoch_curve_2024-05-30_11.39.png

+ 0
- 0
figures/Pre-Clinical Data Run/cnn_net.pth → figures/Pre-Clinical_Data_Run/cnn_net.pth
+ 0
- 0
figures/Pre-Clinical_Data_Run/cnn_net_data_2024-05-30_11:39.csv → figures/Pre-Clinical_Data_Run/cnn_net_data_2024-05-30_11.39.csv
+ 0
- 0
figures/Pre-Clinical_Data_Run/confusion_matrix_2024-05-30_11:39.png → figures/Pre-Clinical_Data_Run/confusion_matrix_2024-05-30_11.39.png
+ 0
- 0
figures/ROC_2024-07-19_12:31.png → figures/ROC_2024-07-19_12.31.png
+ 0
- 0
figures/avgloss_epoch_curve_2024-07-19_12:30.png → figures/avgloss_epoch_curve_2024-07-19_12.30.png
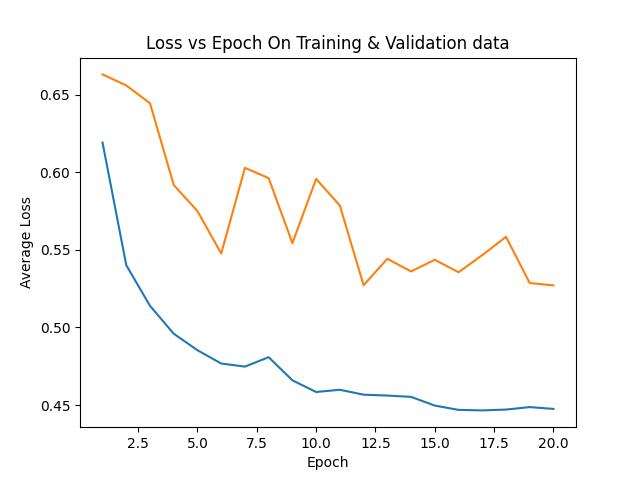
+ 0
- 0
figures/cnn_net_data_2024-07-19_12:30.csv → figures/cnn_net_data_2024-07-19_12.30.csv
+ 0
- 0
figures/confusion_matrix_2024-07-19_12:31.png → figures/confusion_matrix_2024-07-19_12.31.png
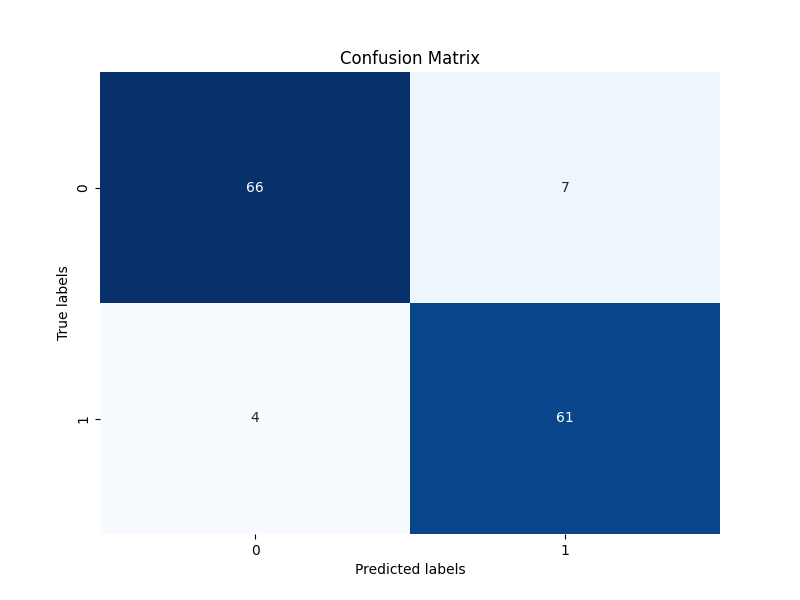
+ 36
- 9
main.py
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
+ 1
- 1
original_model/innvestigate/analyzer/__init__.py
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
+ 15
- 7
utils/CNN.py
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
+ 5
- 3
utils/dataset_sd_mean_finder.py
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
+ 6
- 3
utils/preprocess.py
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
+ 8
- 6
utils/train_methods.py
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||