|
|
@@ -1,516 +0,0 @@
|
|
|
-import slicer
|
|
|
-import os
|
|
|
-import subprocess
|
|
|
-import re
|
|
|
-import slicerNetwork
|
|
|
-import ctk,qt
|
|
|
-import json
|
|
|
-
|
|
|
-dicomModify=os.getenv("HOME")
|
|
|
-if not dicomModify==None:
|
|
|
- dicomModify+="/software/install/"
|
|
|
- dicomModify+="dicomModify/bin/dicomModify"
|
|
|
-
|
|
|
-class loadDicom(slicer.ScriptedLoadableModule.ScriptedLoadableModule):
|
|
|
- def __init__(self,parent):
|
|
|
- slicer.ScriptedLoadableModule.ScriptedLoadableModule.__init__(self, parent)
|
|
|
- self.className="loadDicom"
|
|
|
- self.parent.title="loadDicom"
|
|
|
- self.parent.categories = ["LabKey"]
|
|
|
- self.parent.dependencies = []
|
|
|
- self.parent.contributors = ["Andrej Studen (UL/FMF)"] # replace with "Firstname Lastname (Organization)"
|
|
|
- self.parent.helpText = """
|
|
|
- utilities for parsing dicom entries
|
|
|
- """
|
|
|
- self.parent.acknowledgementText = """
|
|
|
- Developed within the medical physics research programme of the Slovenian research agency.
|
|
|
- """ # replace with organization, grant and thanks.
|
|
|
-
|
|
|
-class loadDicomWidget(slicer.ScriptedLoadableModule.ScriptedLoadableModuleWidget):
|
|
|
- """Uses ScriptedLoadableModuleWidget base class, available at:
|
|
|
- https://github.com/Slicer/Slicer/blob/master/Base/Python/slicer/ScriptedLoadableModule.py
|
|
|
- """
|
|
|
-
|
|
|
- def setup(self):
|
|
|
- slicer.ScriptedLoadableModule.ScriptedLoadableModuleWidget.setup(self)
|
|
|
- self.logic=loadDicomLogic(self)
|
|
|
- self.network=slicerNetwork.labkeyURIHandler()
|
|
|
-
|
|
|
- connectionCollapsibleButton = ctk.ctkCollapsibleButton()
|
|
|
- connectionCollapsibleButton.text = "Connection"
|
|
|
- self.layout.addWidget(connectionCollapsibleButton)
|
|
|
-
|
|
|
- connectionFormLayout = qt.QFormLayout(connectionCollapsibleButton)
|
|
|
-
|
|
|
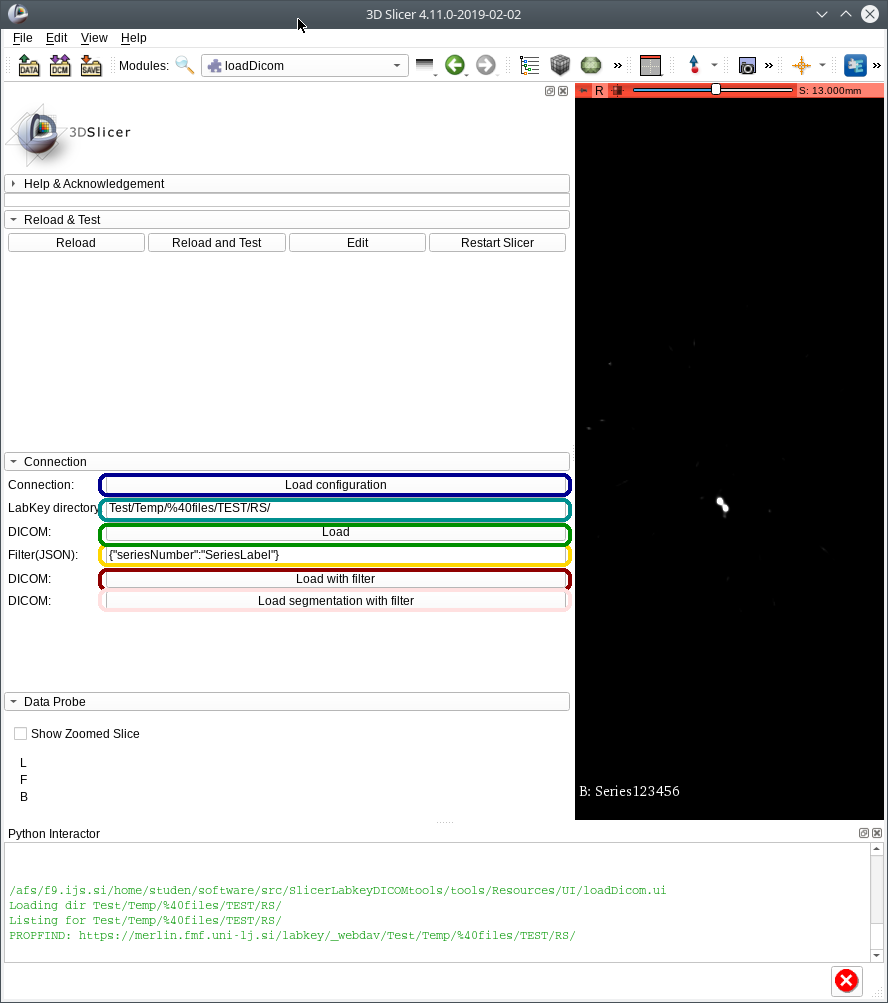
- self.loadConfigButton=qt.QPushButton("Load configuration")
|
|
|
- self.loadConfigButton.toolTip="Load configuration"
|
|
|
- self.loadConfigButton.connect('clicked(bool)',self.onLoadConfigButtonClicked)
|
|
|
- connectionFormLayout.addRow("Connection:",self.loadConfigButton)
|
|
|
-
|
|
|
- self.DICOMDirectory=qt.QLineEdit("Test/Temp/%40files/TEST/MLEM")
|
|
|
- connectionFormLayout.addRow("LabKey directory:",self.DICOMDirectory)
|
|
|
-
|
|
|
-
|
|
|
- loadDICOMButton=qt.QPushButton("Load")
|
|
|
- loadDICOMButton.toolTip="Load DICOM"
|
|
|
- loadDICOMButton.clicked.connect(self.onLoadDICOMButtonClicked)
|
|
|
- connectionFormLayout.addRow("DICOM:",loadDICOMButton)
|
|
|
-
|
|
|
- self.DICOMFilter=qt.QLineEdit('{"seriesNumber":"SeriesLabel"}')
|
|
|
- connectionFormLayout.addRow("Filter(JSON):",self.DICOMFilter)
|
|
|
-
|
|
|
- loadDICOMFilterButton=qt.QPushButton("Load with filter")
|
|
|
- loadDICOMFilterButton.toolTip="Load DICOM with filter"
|
|
|
- loadDICOMFilterButton.clicked.connect(self.onLoadDICOMFilterButtonClicked)
|
|
|
- connectionFormLayout.addRow("DICOM:",loadDICOMFilterButton)
|
|
|
-
|
|
|
- loadDICOMSegmentationFilterButton=qt.QPushButton("Load segmentation with filter")
|
|
|
- loadDICOMSegmentationFilterButton.toolTip="Load DICOM (RT contour) with filter"
|
|
|
- loadDICOMSegmentationFilterButton.clicked.connect(self.onLoadDICOMSegmentationFilterButtonClicked)
|
|
|
- connectionFormLayout.addRow("DICOM:",loadDICOMSegmentationFilterButton)
|
|
|
-
|
|
|
-
|
|
|
- def onLoadConfigButtonClicked(self):
|
|
|
- filename=qt.QFileDialog.getOpenFileName(None,'Open configuration file (JSON)',
|
|
|
- os.path.join(os.path.expanduser('~'),'.labkey'), '*.json')
|
|
|
- self.network.parseConfig(filename)
|
|
|
- self.network.initRemote()
|
|
|
- self.loadConfigButton.setText(os.path.basename(filename))
|
|
|
-
|
|
|
- def onLoadDICOMFilterButtonClicked(self):
|
|
|
- filter=json.loads(self.DICOMFilter.text)
|
|
|
- #print("Filter is {}".format(filter))
|
|
|
- self.logic.loadVolumes(self.network,self.DICOMDirectory.text,filter)
|
|
|
-
|
|
|
- def onLoadDICOMSegmentationFilterButtonClicked(self):
|
|
|
- filter=json.loads(self.DICOMFilter.text)
|
|
|
- #print("Filter is {}".format(filter))
|
|
|
- self.logic.loadSegmentations(self.network,self.DICOMDirectory.text,filter)
|
|
|
-
|
|
|
- def onLoadDICOMButtonClicked(self):
|
|
|
- self.logic.load(self.network,self.DICOMDirectory.text)
|
|
|
-
|
|
|
-
|
|
|
-class loadDicomLogic(slicer.ScriptedLoadableModule.ScriptedLoadableModuleLogic):
|
|
|
- def __init__(self,parent):
|
|
|
- slicer.ScriptedLoadableModule.ScriptedLoadableModuleLogic.__init__(self, parent)
|
|
|
-
|
|
|
- self.tag={
|
|
|
- 'sopInstanceUid' : {'tag':"0008,0018",'VR':'UI'},
|
|
|
- 'studyDate': {'tag':"0008,0020",'VR':'DA'},
|
|
|
- 'studyTime': {'tag':"0008,0030",'VR':'TM'},
|
|
|
- 'seriesTime': {'tag':"0008,0031",'VR':'TM'},
|
|
|
- 'modality': {'tag':"0008,0060",'VR':'CS'},
|
|
|
- 'presentationIntentType': {'tag':"0008,0068",'VR':'CS'},
|
|
|
- 'manufacturer': {'tag':"0008,0070",'VR':'LO'},
|
|
|
- 'institutionName': {'tag':"0008,0080",'VR':'LO'},
|
|
|
- 'studyDescription': {'tag':"0008,1030",'VR':'LO'},
|
|
|
- 'seriesDescription': {'tag':"0008,103e",'VR':'LO'},
|
|
|
- 'manufacturerModelName': {'tag':"0008,1090",'VR':'LO'},
|
|
|
- 'patientName': {'tag':"0010,0010",'VR':'PN'},
|
|
|
- 'patientId': {'tag':"0010,0020",'VR':'LO'},
|
|
|
- 'patientBirthDate': {'tag':"0010,0030",'VR':'DA'},
|
|
|
- 'patientSex': {'tag':"0010,0040",'VR':'CS'},
|
|
|
- 'patientAge': {'tag':"0010,1010",'VR':'AS'},
|
|
|
- 'patientComments': {'tag':"0010,4000",'VR':'LT'},
|
|
|
- 'sequenceName': {'tag':"0018,0024",'VR':'SH'},
|
|
|
- 'kVP': {'tag':"0018,0060",'VR':'DS'},
|
|
|
- 'percentPhaseFieldOfView': {'tag':"0018,0094",'VR':'DS'},
|
|
|
- 'xRayTubeCurrent': {'tag':"0018,1151",'VR':'IS'},
|
|
|
- 'exposure': {'tag':"0018,1152",'VR':'IS'},
|
|
|
- 'imagerPixelSpacing': {'tag':"0018,1164",'VR':'DS'},
|
|
|
- 'bodyPartThickness': {'tag':"0018,11a0",'VR':'DS'},
|
|
|
- 'compressionForce': {'tag':"0018,11a2",'VR':'DS'},
|
|
|
- 'viewPosition': {'tag':"0018,5101",'VR':'CS'},
|
|
|
- 'fieldOfViewHorizontalFlip': {'tag':"0018,7034",'VR':'CS'},
|
|
|
- 'studyInstanceUid': {'tag':"0020,000d",'VR':'UI'},
|
|
|
- 'seriesInstanceUid': {'tag':"0020,000e",'VR':'UI'},
|
|
|
- 'studyId': {'tag':"0020,0010",'VR':'SH'},
|
|
|
- 'seriesNumber': {'tag':"0020,0011",'VR':'IS'},
|
|
|
- 'instanceNumber': {'tag':"0020,0013",'VR':'IS'},
|
|
|
- 'frameOfReferenceInstanceUid': {'tag':"0020,0052",'seqTag':"3006,0010",'VR':'UI'},
|
|
|
- 'imageLaterality': {'tag':"0020,0062",'VR':'CS'},
|
|
|
- 'imagesInAcquisition': {'tag':"0020,1002",'VR':'IS'},
|
|
|
- 'photometricInterpretation': {'tag':"0028,0004",'VR':'CS'},
|
|
|
- 'reconstructionMethod': {'tag':"0054,1103",'VR':'LO'}
|
|
|
- }
|
|
|
- self.tagPyDicom={
|
|
|
- 'studyDate': 0x00080020,
|
|
|
- 'studyTime': 0x00080030,
|
|
|
- 'modality': 0x00080060,
|
|
|
- 'presentationIntentType': 0x00080068,
|
|
|
- 'manufacturer': 0x00080070,
|
|
|
- 'studyDescription': 0x00081030,
|
|
|
- 'seriesDescription': 0x0008103e,
|
|
|
- 'patientName': 0x00100010,
|
|
|
- 'patientId': 0x00100020,
|
|
|
- 'patientBirthDate': 0x00100030,
|
|
|
- 'patientSex': 0x00100040,
|
|
|
- 'patientComments': 0x00104000,
|
|
|
- 'sequenceName': 0x00180024,
|
|
|
- 'kVP': 0x00180060,
|
|
|
- 'percentPhaseFieldOfView': 0x00180094,
|
|
|
- 'xRayTubeCurrent': 0x00181151,
|
|
|
- 'exposure': 0x00181152,
|
|
|
- 'imagerPixelSpacing': 0x00181164,
|
|
|
- 'bodyPartThickness': 0x001811a0,
|
|
|
- 'compressionForce': 0x001811a2,
|
|
|
- 'viewPosition': 0x00185101,
|
|
|
- 'studyInstanceUid': 0x0020000d,
|
|
|
- 'seriesInstanceUid': 0x0020000e,
|
|
|
- 'studyId': 0x00200010,
|
|
|
- 'seriesNumber': 0x00200011,
|
|
|
- 'instanceNumber': 0x00200013,
|
|
|
- 'frameOfReferenceInstanceUid': 0x00200052
|
|
|
- }
|
|
|
- #new_dict_items={
|
|
|
- # 0x001811a0: ('DS','BodyPartThickness','Body Part Thickness')
|
|
|
- #}
|
|
|
- #dicom.datadict.add_dict_entries(new_dict_items)
|
|
|
- self.local=False
|
|
|
- self.useDicomModify=False
|
|
|
-
|
|
|
- def enableDicomModify(self):
|
|
|
- self.useDicomModify=True
|
|
|
-
|
|
|
- def setLocal(self,basePath):
|
|
|
- self.local=True
|
|
|
- self.basePath=basePath
|
|
|
-
|
|
|
-
|
|
|
- def getHex(self,key):
|
|
|
- #convert string to hex key;
|
|
|
- fv=key.split(",")
|
|
|
- return int(fv[0],16)*0x10000+int(fv[1],16)
|
|
|
-
|
|
|
- def load(self,sNet,dir,doRemove=True):
|
|
|
- #load directory using DICOMLib tools
|
|
|
-
|
|
|
- print("Loading dir {}").format(dir)
|
|
|
- dicomFiles=self.listdir(sNet,dir)
|
|
|
-
|
|
|
- filelist=[]
|
|
|
- for f in dicomFiles:
|
|
|
- localPath=self.getfile(sNet,f)
|
|
|
- filelist.append(localPath)
|
|
|
- if not self.useDicomModify:
|
|
|
- continue
|
|
|
- if dicomModify==None:
|
|
|
- continue
|
|
|
- f0=localPath
|
|
|
- f1=f0+"1"
|
|
|
- try:
|
|
|
- subprocess.call(\
|
|
|
- dicomModify+" "+f0+" "+f1+" && mv "+f1+" "+f0+";",
|
|
|
- shell=True)
|
|
|
- except OSError:
|
|
|
- print("dicomModify failed")
|
|
|
-
|
|
|
- try:
|
|
|
- loadables=self.volumePlugin.examineForImport([filelist])
|
|
|
- except AttributeError:
|
|
|
- self.volumePlugin=slicer.modules.dicomPlugins['DICOMScalarVolumePlugin']()
|
|
|
- loadables=self.volumePlugin.examineForImport([filelist])
|
|
|
-
|
|
|
-
|
|
|
- for loadable in loadables:
|
|
|
- #check if it makes sense to load a particular loadable
|
|
|
-
|
|
|
- if loadable.name.find('imageOrientationPatient')>-1:
|
|
|
- continue
|
|
|
- filter={}
|
|
|
- filter['seriesNumber']=None
|
|
|
- metadata={}
|
|
|
- if not self.applyFilter(loadable,filter,metadata):
|
|
|
- continue
|
|
|
- volumeNode=self.volumePlugin.load(loadable)
|
|
|
- if volumeNode != None:
|
|
|
- vName='Series'+metadata['seriesNumber']
|
|
|
- volumeNode.SetName(vName)
|
|
|
-
|
|
|
- try:
|
|
|
- loadableRTs=self.RTPlugin.examineForImport([filelist])
|
|
|
- except:
|
|
|
- self.RTPlugin=plugin=slicer.modules.dicomPlugins['DicomRtImportExportPlugin']()
|
|
|
- loadableRTs=self.RTPlugin.examineForImport([filelist])
|
|
|
-
|
|
|
- for loadable in loadableRTs:
|
|
|
- segmentationNode=self.RTPlugin.load(loadable)
|
|
|
-
|
|
|
- if not doRemove:
|
|
|
- return
|
|
|
-
|
|
|
- for f in filelist:
|
|
|
- self.removeLocal(f)
|
|
|
-
|
|
|
- def applyFilter(self,loadable,filter,nodeMetadata):
|
|
|
- #apply filter to loadable.file[0]. Return true if file matches prescribed filter and
|
|
|
- #false otherwise
|
|
|
-
|
|
|
- #filter is a directory with keys equal to pre-specified values listed above
|
|
|
- #if value associated to key equals None, that value gets set in nodeMetadata
|
|
|
- #if value is set, a match is attempted and result reported in return value
|
|
|
- #all filters should match for true output
|
|
|
-
|
|
|
- return self.applyFilterFile(loadable.files[0],filter,nodeMetadata)
|
|
|
-
|
|
|
- def applyFilterFile(self,file,filter,nodeMetadata,shell=False):
|
|
|
- #apply filter to file. Return true if file matches prescribed filter and
|
|
|
- #false otherwise
|
|
|
-
|
|
|
- #filter is a directory with keys equal to pre-specified values listed above
|
|
|
- #if value associated to key equals None, that value gets set in nodeMetadata
|
|
|
- #if value is set, a match is attempted and result reported in return value
|
|
|
- #all filters should match for true output
|
|
|
-
|
|
|
- filterOK=True
|
|
|
-
|
|
|
- for key in filter:
|
|
|
- try:
|
|
|
- fileValue=dicomValue(file,self.tag[key]['tag'],self.tag[key]['seqTag'],shell=shell)
|
|
|
- except KeyError:
|
|
|
- fileValue=dicomValue(file,self.tag[key]['tag'],shell=shell)
|
|
|
-
|
|
|
-
|
|
|
- if filter[key]=="SeriesLabel":
|
|
|
- nodeMetadata['seriesLabel']=fileValue
|
|
|
- continue
|
|
|
-
|
|
|
- if not filter[key]==None:
|
|
|
- if not fileValue==filter[key]:
|
|
|
- #print("File {} failed for tag {}: {}/{}").format(\
|
|
|
- # file,key,fileValue,filter[key])
|
|
|
- filterOK=False
|
|
|
- break
|
|
|
-
|
|
|
- nodeMetadata[key]=fileValue
|
|
|
-
|
|
|
-
|
|
|
- return filterOK
|
|
|
-
|
|
|
- def listdir(self,sNet,dir):
|
|
|
- #list remote directory
|
|
|
- if self.local:
|
|
|
- dir1=os.path.join(self.basePath,dir)
|
|
|
- dirs=os.listdir(dir1)
|
|
|
- return [os.path.join(dir1,f) for f in dirs]
|
|
|
- return sNet.listRelativeDir(dir)
|
|
|
-
|
|
|
- def getfile(self,sNet,file):
|
|
|
- #get remote file
|
|
|
- if self.local:
|
|
|
- return file
|
|
|
- return sNet.DownloadFileToCache(file)
|
|
|
-
|
|
|
- def removeLocal(self,localFile):
|
|
|
- if self.local:
|
|
|
- return
|
|
|
- os.remove(localFile)
|
|
|
-
|
|
|
-
|
|
|
- def loadVolumes(self,sNet,dir,filter,doRemove=True):
|
|
|
- #returns all series from the directory, each as a separate node in a node list
|
|
|
- #filter is a dictionary of speciifed dicom values, if filter(key)=None, that values
|
|
|
- #get set, if it isn't, the file gets checked for a match
|
|
|
-
|
|
|
- print("Loading dir {}").format(dir)
|
|
|
- dicomFiles=self.listdir(sNet,dir)
|
|
|
- #filelist=[os.path.join(dir,f) for f in os.listdir(dir)]
|
|
|
- filelist=[]
|
|
|
- for f in dicomFiles:
|
|
|
- localPath=self.getfile(sNet,f)
|
|
|
- f0=localPath
|
|
|
- f1=f0+"1"
|
|
|
- if not dicomModify==None:
|
|
|
- try:
|
|
|
- subprocess.call(dicomModify+" "+f0+" "+f1+" && mv "+f1+" "+f0+";", shell=False)
|
|
|
- except OSError:
|
|
|
- print("dicomModify failed")
|
|
|
- filelist.append(localPath)
|
|
|
-
|
|
|
- try:
|
|
|
- loadables=self.volumePlugin.examineForImport([filelist])
|
|
|
- except AttributeError:
|
|
|
- self.volumePlugin=slicer.modules.dicomPlugins['DICOMScalarVolumePlugin']()
|
|
|
- loadables=self.volumePlugin.examineForImport([filelist])
|
|
|
-
|
|
|
-
|
|
|
- volumeNodes=[]
|
|
|
- print("Number of loadables:{}").format(len(loadables))
|
|
|
- for loadable in loadables:
|
|
|
- #TODO check if it makes sense to load a particular loadable
|
|
|
-
|
|
|
- print("{}: Checking number of files: {}".format(\
|
|
|
- loadable.name,len(loadable.files)))
|
|
|
-
|
|
|
- #perform checks
|
|
|
- fileMetadata={}
|
|
|
- loadable.files[:]=[f for f in loadable.files \
|
|
|
- if self.applyFilterFile(f,filter,fileMetadata)]
|
|
|
-
|
|
|
- if len(loadable.files)<1:
|
|
|
- #skip this loadable
|
|
|
- continue
|
|
|
-
|
|
|
- print("{}: Final number of files: {}".format(\
|
|
|
- loadable.name,len(loadable.files)))
|
|
|
-
|
|
|
- nodeMetadata={}
|
|
|
- self.applyFilterFile(loadable.files[0],filter,nodeMetadata)
|
|
|
-
|
|
|
-
|
|
|
-
|
|
|
- volumeNode=self.volumePlugin.load(loadable,"DCMTK")
|
|
|
- if volumeNode != None:
|
|
|
- vName='Series'+nodeMetadata['seriesLabel']
|
|
|
- volumeNode.SetName(vName)
|
|
|
- volume={'node':volumeNode,'metadata':nodeMetadata}
|
|
|
- volumeNodes.append(volume)
|
|
|
-
|
|
|
- if self.local:
|
|
|
- return volumeNodes
|
|
|
-
|
|
|
- if doRemove:
|
|
|
- for f in filelist:
|
|
|
- self.removeLocal(f)
|
|
|
-
|
|
|
- return volumeNodes
|
|
|
-
|
|
|
- def loadSegmentations(self,net,dir,filter,doRemove=True):
|
|
|
-
|
|
|
- print("Loading dir {}").format(dir)
|
|
|
- dicomFiles=self.listdir(net,dir)
|
|
|
- filelist=[self.getfile(net,f) for f in dicomFiles]
|
|
|
- segmentationNodes=[]
|
|
|
-
|
|
|
- try:
|
|
|
- loadableRTs=self.RTPlugin.examineForImport([filelist])
|
|
|
- except:
|
|
|
- self.RTPlugin=plugin=slicer.modules.dicomPlugins['DicomRtImportExportPlugin']()
|
|
|
- loadableRTs=self.RTPlugin.examineForImport([filelist])
|
|
|
-
|
|
|
- for loadable in loadableRTs:
|
|
|
-
|
|
|
- nodeMetadata={}
|
|
|
- filterOK=self.applyFilter(loadable,filter,nodeMetadata)
|
|
|
-
|
|
|
- if not filterOK:
|
|
|
- continue
|
|
|
-
|
|
|
- success=self.RTPlugin.load(loadable)
|
|
|
-
|
|
|
- if not success:
|
|
|
- print("Could not load RT structure set")
|
|
|
- return
|
|
|
-
|
|
|
- segNodes=slicer.util.getNodesByClass("vtkMRMLSegmentationNode")
|
|
|
- segmentationNode=segNodes[0]
|
|
|
- #assume we loaded the first node in list
|
|
|
-
|
|
|
- if segmentationNode != None:
|
|
|
- sName='Segmentation'+nodeMetadata['seriesLabel']
|
|
|
- segmentationNode.SetName(sName)
|
|
|
- segmentation={'node':segmentationNode,'metadata':nodeMetadata}
|
|
|
- segmentationNodes.append(segmentation)
|
|
|
-
|
|
|
- if not doRemove:
|
|
|
- return segmentationNodes
|
|
|
-
|
|
|
- for f in filelist:
|
|
|
- self.removeLocal(f)
|
|
|
-
|
|
|
- return segmentationNodes
|
|
|
-
|
|
|
-def isDicom(file):
|
|
|
- #check if file is a dicom file
|
|
|
- try:
|
|
|
- f=open(file,'rb')
|
|
|
- except IOError:
|
|
|
- return False
|
|
|
-
|
|
|
- f.read(128)
|
|
|
- dt=f.read(4)
|
|
|
- f.close()
|
|
|
- return dt=='DICM'
|
|
|
-
|
|
|
-
|
|
|
-def dicomValue(fileName,tag,seqTag=None,shell=False):
|
|
|
- #query dicom value of file using dcmdump (DCMTK routine)
|
|
|
-
|
|
|
- #shell must be false for *nix
|
|
|
- if os.name=="posix":
|
|
|
- shell=False
|
|
|
-
|
|
|
- debug=False
|
|
|
- dcmdump=os.path.join(os.environ['SLICER_HOME'],"bin","dcmdump")
|
|
|
-
|
|
|
- try:
|
|
|
- if debug:
|
|
|
- print("Calling {}".format(dcmdump))
|
|
|
- out=subprocess.check_output([dcmdump,'+p','+P',tag,fileName],shell=shell)
|
|
|
- if debug:
|
|
|
- print("Got {}".format(out))
|
|
|
- except subprocess.CalledProcessError as e:
|
|
|
- return None
|
|
|
-
|
|
|
-
|
|
|
- if debug:
|
|
|
- print("Tag {} Line '{}'").format(tag,out)
|
|
|
- if len(out)==0:
|
|
|
- return out
|
|
|
-
|
|
|
- #parse multi-match outputs which appear as several lines
|
|
|
- lst=out.split('\n')
|
|
|
- return getTagValue(lst,tag,seqTag)
|
|
|
-
|
|
|
-def getTagValue(lst,tag,seqTag=None):
|
|
|
- #report tag value from a list lst of lines reported by dcmdump
|
|
|
-
|
|
|
- debug=False
|
|
|
-
|
|
|
- #parse output
|
|
|
- longTag="^\({}\)".format(tag)
|
|
|
- #combine sequence and actual tags to long tags
|
|
|
- if not seqTag==None:
|
|
|
- if debug:
|
|
|
- print("Tag:{} seqTag:{}").format(tag,seqTag)
|
|
|
- longTag="^\({}\).\({}\)".format(seqTag,tag)
|
|
|
-
|
|
|
- #pick the values
|
|
|
- pattern=r'^.*\[(.*)\].*$'
|
|
|
-
|
|
|
- #extract tag values
|
|
|
- rpl=[re.sub(pattern,r'\1',f) for f in lst]
|
|
|
-
|
|
|
- #logical whether the line can be matched to typical dcmdump output
|
|
|
- mtchPattern=[re.match(pattern,f) for f in lst]
|
|
|
- #find matching tags
|
|
|
- mtchTag=[re.match(longTag,f) for f in lst]
|
|
|
-
|
|
|
- #weed out non-matching lines and lines not matching longTag
|
|
|
- mtch=[None if x==None or y==None else x \
|
|
|
- for x,y in zip(mtchTag,mtchPattern)]
|
|
|
- #set values
|
|
|
- out=[x for y,x in zip(mtch,rpl) if not y==None]
|
|
|
- if len(out)==0:
|
|
|
- return ''
|
|
|
- #return first match
|
|
|
- out=out[0]
|
|
|
-
|
|
|
- if debug:
|
|
|
- print("Tag {} Parsed value {}").format(tag,out)
|
|
|
- #split output to lists if values are DICOM lists
|
|
|
- if out.find('\\')>-1:
|
|
|
- out=out.split('\\')
|
|
|
-
|
|
|
- return out
|
|
|
-
|
|
|
-
|
|
|
-def clearNodes():
|
|
|
- nodes=[]
|
|
|
- nodes.extend(slicer.util.getNodesByClass("vtkMRMLScalarVolumeNode"))
|
|
|
- nodes.extend(slicer.util.getNodesByClass("vtkMRMLScalarVolumeDisplayNode"))
|
|
|
- nodes.extend(slicer.util.getNodesByClass("vtkMRMLSegmentationNode"))
|
|
|
- nodes.extend(slicer.util.getNodesByClass("vtkMRMLSegmentationDisplayNode"))
|
|
|
- for node in nodes:
|
|
|
- slicer.mrmlScene.RemoveNode(node)
|